DNA Interactions in Crowded Nanopores
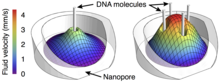
The motion of DNA in crowded environments is a common theme in physics and biology. Examples include gel electrophoresis and the self-interaction of DNA within cells and viral capsids. Here we study the interaction of multiple DNA molecules within a nanopore by tethering the DNA to a bead held in a laser optical trap to produce a "molecular tug-of-war". We measure this tether force as a function of the number of DNA molecules in the pore and show that the force per molecule decreases with the number of molecules. A scaling argument combined with numerical simulations suggested that the hydrodynamic interactions between multiple DNA strands explains our observations. At high salt concentrations, when the Debye length approaches the size of the counterions, the force per molecule becomes essentially independent of the number of molecules. We attribute this to a sharp decrease in electroosmotic flow which makes the hydrodynamic interactions ineffective.
ダウンロード
- misiunas_poster.pdf - 3.15MB
- misiunas_abstract.pdf - 0.4MB



