Buoyancy–driven μPCR for DNA Amplification
Application ID: 17227
Polymerase chain reaction (PCR) is one of the most effective methods in molecular biology, medical diagnostics, and biochemical engineering in amplifying a specific sequence of DNA. There has been a great interest in developing portable PCR-based lab-on-a-chip systems for point-of-care applications and one strategy that seems very promising is natural convection-based PCR.
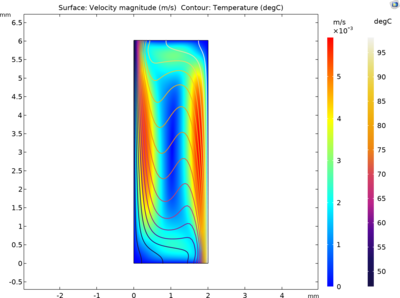
This model studies the spatio-temporal variation of the concentrations of single-stranded (ssDNA), double-stranded (dsDNA) and primer-annealed (aDNA) DNA components. The driving mechanism for the fluid motion is the temperature-induced density differences that results in buoyancy flow. This approach eliminates the need for an external driving mechanism and the difference between the temperatures is sufficient to circulate the PCR mixture and amplify DNA in a closed loop.
この model の例は, 通常次の製品を使用して構築されるこのタイプのアプリケーションを示しています.
ただし, これを完全に定義およびモデル化するには, 追加の製品が必要になる場合があります. さらに, この例は, 次の製品の組み合わせのコンポーネントを使用して定義およびモデル化することもできます.
アプリケーションのモデリングに必要な COMSOL® 製品の組み合わせは, 境界条件, 材料特性, フィジックスインターフェース, パーツライブラリなど, いくつかの要因によって異なります. 特定の機能が複数の製品に共通している場合もあります. お客様のモデリングニーズに適した製品の組み合わせを決定するために, 製品仕様一覧 を確認し, 無償のトライアルライセンスをご利用ください. COMSOL セールスおよびサポートチームでは, この件に関するご質問にお答えしています.
